import plotly.graph_objects as go
import plotly.io as pio
import plotly.express as px
from IPython.display import IFrame
# Retrieve creator names and ORCID ids from all publications
all_creator_ids = []
all_creator_ids_set = set([])
creator_id2name = {}
publications = data['person']['publications']
for r in publications['nodes']:
if r['versionOfCount'] > 0:
# If the current output is a version of another one, exclude it
continue
creator_ids = list(filter(None, [s['id'] for s in r['creators']]))
all_creator_ids_set.update(creator_ids)
all_creator_ids.append(creator_ids)
for creator in r['creators']:
if (creator['id'] not in creator_id2name and creator['id'] is not None):
creator_id2name[creator['id']] = creator['name']
# Collect creator names into all_unique_creator_names - these will be labels in the sankey plot
# Initialise coauthorship_matrix, that will be used to populate lists needed for the sankey plot
all_unique_creator_ids = list(all_creator_ids_set)
length = len(all_unique_creator_ids)
coauthorship_matrix = []
all_unique_creator_names = []
for id in all_unique_creator_ids:
all_unique_creator_names.append(creator_id2name[id])
coauthorship_matrix.append([0] * length)
# Populate coauthorship_matrix
for cids in all_creator_ids:
for cid in cids:
c_pos = all_unique_creator_ids.index(cid)
for cid in cids:
co_pos = all_unique_creator_ids.index(cid)
if c_pos != co_pos:
coauthorship_matrix[c_pos][co_pos] += 1
# Use coauthorship_matrix to populate lists needed for the sankey diagram: sourceIndexes, targetIndexes and linkWeights
# For Plotly colour swatches, see: https://plotly.com/python/builtin-colorscales/
colRange = px.colors.sequential.matter;
maxColIndex = len(colRange)
sourceIndexes = []
targetIndexes = []
linkWeights = []
linkColours = []
for c_pos, r in enumerate(coauthorship_matrix):
# On the left hand side of sankey retain only the researcher in question
if all_unique_creator_ids[c_pos] != query_params['researcherId']:
continue
for co_pos, weight in enumerate(r):
if coauthorship_matrix[c_pos][co_pos] > 1:
# Include links to co-authors of at least 2 publications
sourceIndexes.append(c_pos)
targetIndexes.append(co_pos)
linkWeights.append(weight)
linkColours.append(colRange[min(maxColIndex, weight)])
# Create a sankey plot
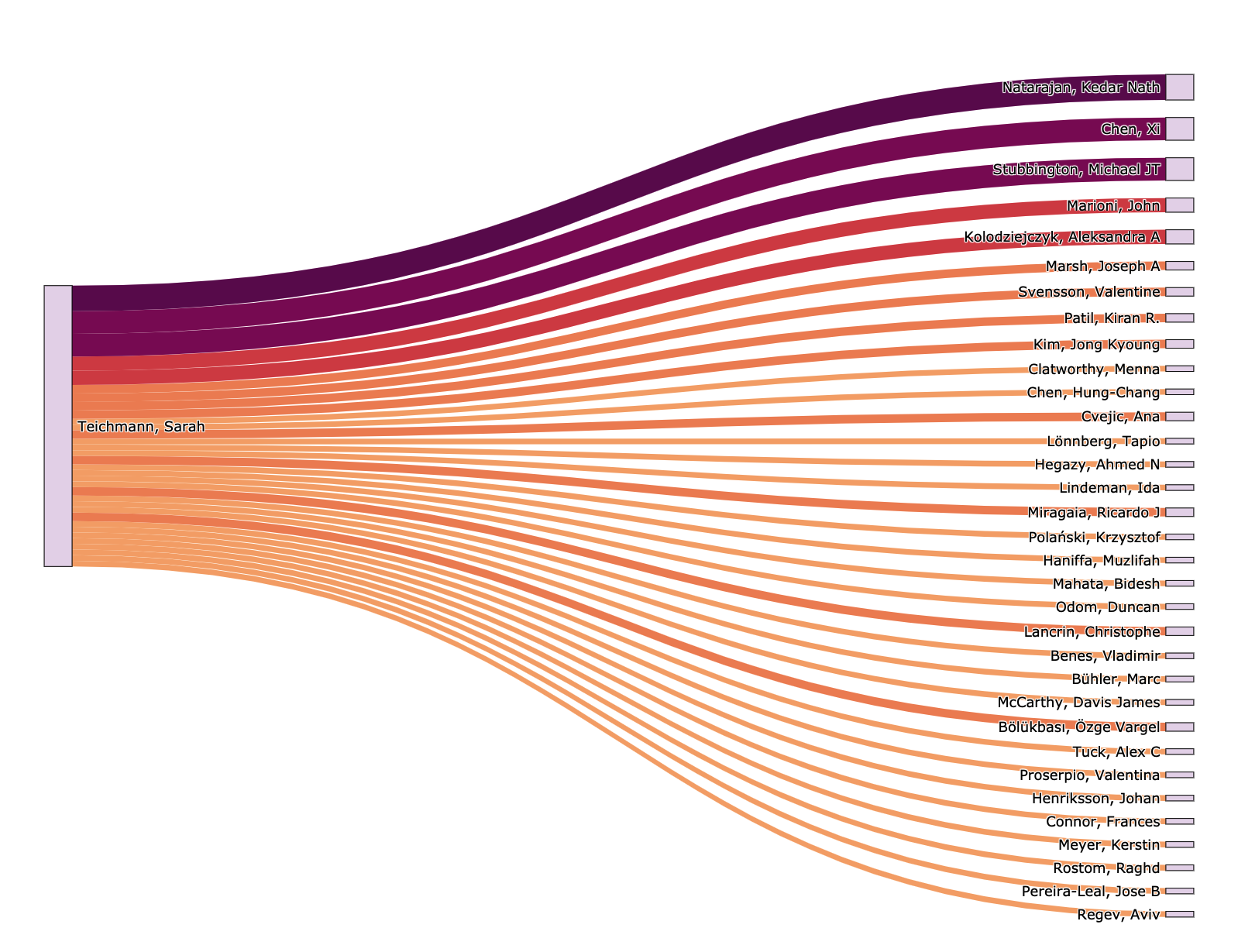
fig = go.Figure(data=[go.Sankey(
node = dict(
pad = 15,
thickness = 20,
line = dict(color = "black", width = 0.5),
label = all_unique_creator_names,
color = "rgba(136,65,157, 0.6)"
),
link = dict(
source = sourceIndexes, # indices correspond to labels in all_unique_creator_names
target = targetIndexes, # ditto
value = linkWeights,
color = linkColours
))])
fig.update_layout(title_text="", font_size=10)
# Write interactive plot out to html file
# pio.write_html(fig, file='out.html')
# Display plot from the saved html file
display(Markdown("### [%s](%s)'s first degree co-authors:" % (creator_id2name[query_params['researcherId']], query_params['researcherId'])))
# IFrame(src="./out.html", width=1000, height=800)
fig.show()