 |
FREYA WP2 User Story 1 | As a data center, I want to see the citations of publications that use my repository for the underlying data, so that I can demonstrate the impact of our repository. |
|---|---|---|
It is important for repositories of scientific data to monitor and report on the impact of the data they store. One useful proxy of that impact are citations of publications accompanying the deposited data.
This notebook uses the DataCite GraphQL API to retrieve data (a.k.a. works) and their citations from three different repositories: PANGAEA, DRYAD and Global Biodiversity Information Facility, using polarstern, butterfly and Lake Malawi as example queries respectively.Goal: By the end of this notebook you should be able to:
- Retrieve works for a chosen repository and query, along with associated metrics such as citation, view and download counts;
- Visualise the work counts over time, e.g.
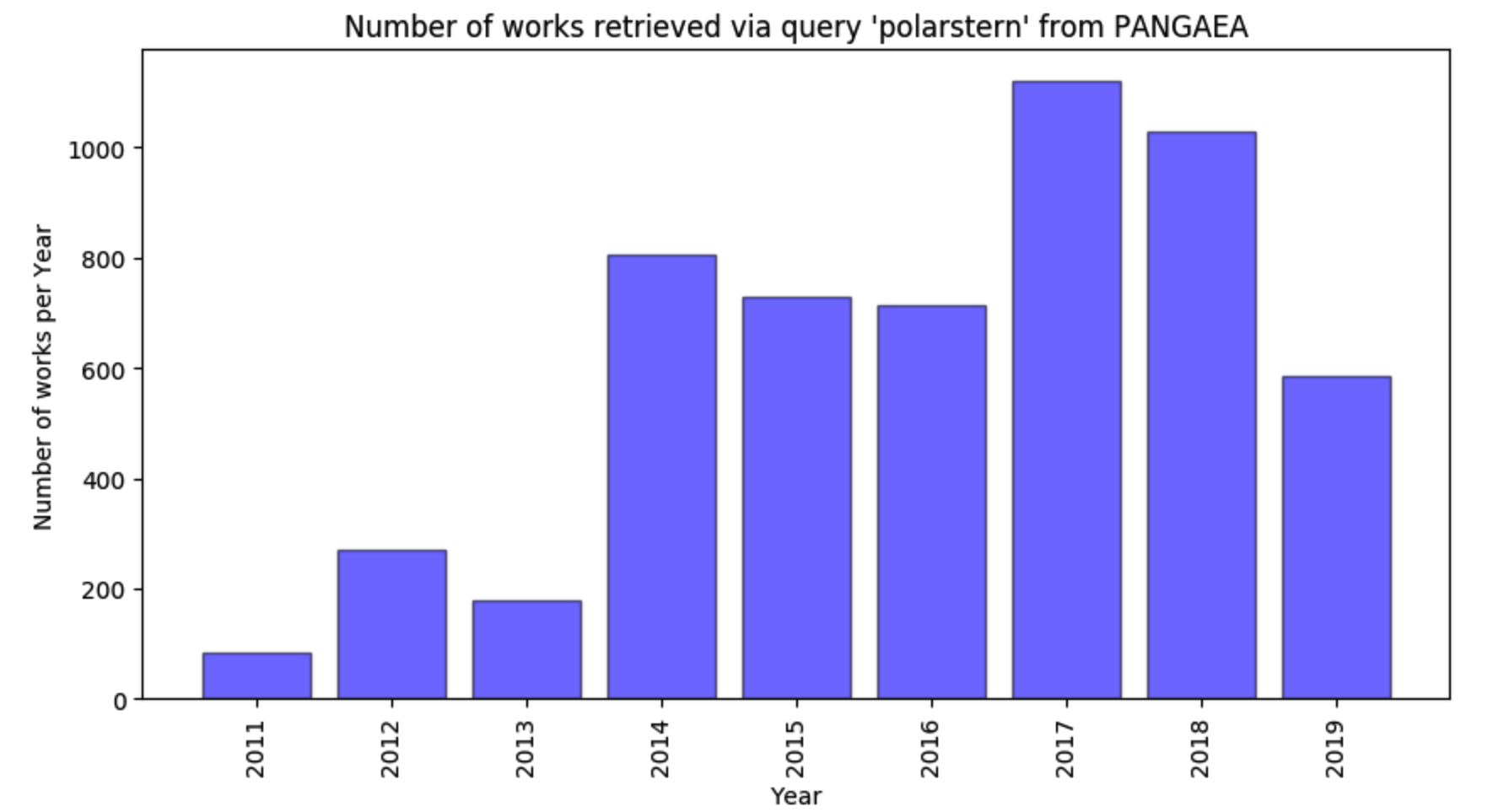
- Present the works in a tabular format and download them in a single BibTeX file;
- For a given work, retrieve all the citations, present them in a tabular format and then download them in a single BibTeX file.